-
纤维素是植物细胞壁的主要多糖类组成成分[1]。根据细胞壁的组成成分和形成方式,可将植物细胞壁分为初生细胞壁和次生细胞壁[1]。初生细胞壁的主要成分是纤维素、半纤维素和果胶,通常在细胞板形成后开始沉积。木本植物木质部细胞在初生细胞壁形成后,进一步分化形成次生细胞壁。次生细胞壁的主要成分是纤维素、半纤维素和木质素,构成了木材的主要成分[2]。纤维素是由D-葡萄糖通过β-1, 4糖苷键连接而成的多聚物,其分子结构非常简单,但合成机制却极其复杂[3]。高等植物中纤维素的合成是在位于细胞膜上的纤维素合酶复合体(Cellulose Synthesis Complex,CSC)完成的,其中纤维素合酶(Cellulose Synthase/CESA)是CSC的主要组成成分,是参与纤维素合成的关键酶[3]。植物的CESA基因最早从棉花(Gossypium hirsutum)纤维中被克隆得到[4],随后,其他植物中的CESA基因相继被报道。拟南芥(Arabidopsis thaliana)基因组包含10个CESA基因,水稻(Oryza sativa)中含有8个CESA基因[5-6]。CESA蛋白序列高度保守,含有8个跨膜结构域,2个在N端,6个在C端[3]。在第2和第3跨膜域之间存在一个胞质环区,该胞质区里含有一些与底物结合与催化相关的残基,这些残基构成了一个保守的D, D, D, Q/RXXRW(天冬氨酸,天冬氨酸,谷氨酰胺-X-X-精氨酸-色氨酸)结构[3]。
目前的研究结果认为,3种CESA蛋白形成一个有功能的CSC[7]。拟南芥中负责次生壁纤维素合成的CSC是由CESA4、CESA7和CESA8组成,负责初生壁纤维素合成的CSC由CESA1、CESA3和CESA6及其类似基因CESA2/CESA5/CESA9组成[5, 8-14]。双分子荧光互补和酵母双杂实验证实,CESA4可与自身互作,且CESA4、CESA7和CESA8之间可以相互作用[15],而免疫共沉淀、双分子荧光互补和酵母双杂实验证明,初生壁CESA1、CESA3和CESA6自身以及相互之间可以相互作用[11, 16]。随后,Pull-down实验证实所有次生壁纤维素合成相关CESA基因之间存在互作关系[13, 17]。
次生细胞壁是木材的主要来源,其中纤维素含量高达40~50%,因而,参与次生壁纤维素合成的CESA基因将在木材形成中发挥关键作用。通过同源序列比对,毛果杨(Populus trichocarpa)基因组中鉴定出17个CESA基因[18-20],其中,负责次生壁纤维素合成CESA基因有5种(PtCESA4、PtCESA7A、PtCESA7B、PtCESA8A和PtCESA8B),相对于拟南芥(AtCESA4、AtCESA7、和AtCESA8)多出2种CESA基因。由于3种CESA蛋白组成一个有功能的CSC,因此,研究杨树5个次生壁CESA基因的表达模式和作用方式,有助于揭示这5个次生壁CESA基因在木本植物纤维素合成中的工作机理。目前,杨树次生壁CESA基因的表达模式已有部分研究,例如,Naoki等通过启动子驱动的GUS转基因材料研究了次生壁CESA基因的在木质部、韧皮部、根冠等组织的表达模式[21]。另外,Song等曾利用PtCESA7A或PtCESA8B的抗体以分化的木质部材料开展共沉淀研究,对共沉淀获得的复合体进行质谱分析检测到了所有5个杨树次生壁CESA基因,表明杨树5个次生壁CESA基因之间存在一定的互作关系,且均参与木材形成过程中的纤维素合成[22]。然而,目前关于杨树5个次生壁CESA基因以何种形式组成有功能的CSC,是随机组合还是有所偏好,尚无相关的研究。本文通过对杨树5个次生壁CESA基因的表达模式与互作模式展开分析,试图为揭示杨树5个次生壁CESA基因的作用模式提供一些线索。
HTML
-
杂种杨84K(P. alba × P. glandulosa)组培苗用于基因遗传转化。84K组培苗在温度为23~25℃、光照为16/8 h(白天/黑夜)、光照强度为50 μmol·m-2·s-1的条件下培养。克隆获得2 Kb的CESA4、CESA7B、CESA8A与CESA8B基因的启动子区,通过Gateway反应重组到pMDC164载体。所有表达载体通过电击转化法转化入根癌农杆菌菌株GV3101,随后采用叶盘法进行遗传转化[23]。简略描述如下:将叶盘与转化了目的构建的农杆菌(OD600为0.4~0.8)共同孵育10 min,之后在分化培养基(MS+0.5 g·L-1 6-苄氨基腺嘌呤(6-Benzylaminopurine,6-BA)+ 0.05 mg·L-1萘乙酸(1-Naphthaleneacetic acid,NAA))上暗培养3天;暗培养后在添加3 mg·L-1潮霉素与200 mg·L-1特美汀的分化培养基上继续培养,获得生长良好的不定芽;将不定芽转移至添加3 mg·L-1潮霉素与200 mg·L-1特美汀的生根培养基(MS+0.05 mg·L-1 IAA + 0.02 mg·L-1 NAA)中继续培养,直到获得不定根生长良好的转基因植株。转基因植株获得后,通过克隆片段特异引物加载体特异引物进行PCR筛选验证转基因阳性植株。每个基因的转基因阳性植株至少拿到10个独立株系。转基因植株培养4 w后进行GUS染色。荧光素酶互补报告实验中所用烟草材料为本氏烟草(Nicotiana benthamiana),在温度为23~25℃、光照为16/8 h(白天/黑夜)的培养室中培养。播种后生长一个月左右可以用于荧光素酶互补报告实验。
-
从TAIR (http://www.arabidopsis.org/)和Phytozome (http://www.phytozome.net/poplar.php)网站获取拟南芥和杨树的CESA基因家族的序列信息。采用MEGA 5.0软件中基于遗传距离的邻近结合法(Neighbor Joining,NJ)构建进化树。根据拟南芥AtCESA锌指结构和跨膜结构域信息(http://www.uniprot.org/),手工绘制杨树CESA锌指结构和跨膜结构域位置图。依据杨树次生壁CESA基因的基因号(PtCESA4:Potri.002G257900;PtCESA7A:Potri.006G181900;PtCESA7B:Potri.018G103900;PtCESA8A:Potri.011G069600;PtCESA8B:Potri.004G059600)在已报道的杨树幼嫩与成熟的茎、叶和根组织的RNA-seq数据[23]与剥皮再生过程再生组织表达量的基因芯片数据[24]中进行检索,获得基因的表达数据。杨树幼嫩与成熟的茎、叶和根组织的RNA-seq实验方法详见Liu等的方法[23],简略而言,使用RNeasy Plant Mini试剂盒和RNase-free DNase Ⅰ试剂盒(Qiagen)提取杨树幼嫩与成熟的茎、叶和根组织的总RNA,交由公司完成测序。剥皮再生过程再生组织表达量的基因芯片数据的实验方法详见Tang等的方法[24],简略而言,取生长时间一致的毛白杨在同一时间进行茎干的环剥,剥皮后第0、7、10、12、16、18和21 d用无菌刀片刮取树干表面再生组织,利用Affymetrix系统平台进行基因芯片的杂交、洗染和扫描。
-
分别用0.1 mol·L-1NAA,0.1 μmol·L-1 6-BA,10 μmol·L-1赤霉素(Gibberellin,GA3),1 nmol·L-1油菜素内酯(epibrassinolide,BR)和1 μmol·L-1乙烯(ethylene),对组培培养4 w的84K植株进行处理。处理方法为,选取生长状态一致的84K组培苗,去除培养基,将根部置于上述浓度的激素溶液中,在处理0 h、3 h、6 h时取样进行次生壁CESA的表达量分析。取样部位为组培苗的茎。所有的试剂购于Sigma-Aldrich。在qRT-PCR实验中,取激素处理后的84K茎段,使用RNeasy Plant Mini试剂盒和RNase-free DNase Ⅰ试剂盒(Qiagen)提取总RNA。每个样品取2 μg RNA使用SuperScript Ⅲ first-strand synthesis system(Invitrogen)合成cDNA第一链。定量PCR的引物使用Primer 3软件设计,按照SYBR Premix Ex TaqTM Ⅱ试剂盒(TaKaRa)使用说明书准备反应混合液,实时定量PCR反应在7500 Fast Real-Time PCR System(Applied Biosystems)或Roche 480实时荧光定量PCR仪(Roche)上进行。PtACTIN作为内参,一次qRT-PCR实验包含四次技术重复。所有实时定量实验至少重复三次。实验所用qRT-PCR引物序列见表 1。
引物名Primer 引物序列Sequence PtCesA4rtF GAGTTGGAGAAATCATCA PtCesA4rtR GAGAGTTAGTTCCTTCAG PtCesA7ArtF CTTCCATGTGCACCTTTGAAGCCA PtCesA7ArtR TCAGGAGCTCGAGGTTCTATGCTA PtCesA7BrtF AACCACTGCAACCATCTCA PtCesA7BrtR ATGTTCCATGACAGCTCAGG PtCesA8ArtF GTATTCTGGGGCTAAACCTTCG PtCesA8ArtR TCTCGCAGTTCATGTAACTCAAC PtCesA8BrtF AAGGACCAAGCAAACATT PtCesA8BrtR AAGTGGATGTAACGGTAAG Table 1. Primers for qRT-PCR.
-
荧光素酶互补报告体系所用载体由中科院遗传所周奕华老师课题组惠赠。按照文献中操作步骤进行荧光素酶互补报告实验[25],简言之,将克隆得到的次生壁PtCESA蛋白CDS序列通过酶切连接的方法分别构建到包含荧光素酶蛋白N端序列的NLuc载体或C端序列的Cluc载体,构建好的载体通过农杆菌GV3101介导转化本氏烟草叶片,培养3 d后,在叶片上涂布荧光素,利用IndiGO软件检测荧光信号。
-
GUS的组织化学染色按照如下步骤进行[23]:培养4 w的转基因组培苗,在预冷的90%丙酮中固定约2 h。固定结束后,在冰上将组织材料使用GUS染色缓冲液(50 mmol·L-1 sodium phosphate (pH7.0),2 mmol·L-1 potassium ferrocyanide,2 mmol·L-1 potassium ferricyanide,10 mmol·L-1 EDTA,0.2% Triton X-100 (v/v))洗涤3次,再转移至GUS染色液(染色缓冲液添加20% (v/v) methanol和1 mM X-Gluc),于37℃温浴12 h后,使用70%乙醇脱色。对CESA4、CESA7B、CESA8A与CESA8B基因启动子驱动的GUS转基因材料,每一个转基因材料至少选择5个株系进行GUS染色,每个转基因材料的染色至少重复三次,以保证结果的可靠性。
1.1. 植物材料
1.2. 生物信息学分析
1.3. 植株激素处理及qRT-PCR分析
1.4. 荧光素酶互补报告体系
1.5. GUS染色分析
-
杨树5个次生壁CESA基因,包含1个CESA4基因,2个CESA7基因和2个CESA8基因,这些基因的序列与拟南芥次生壁CESA基因的序列相似性很高(图 1A)。基因的蛋白结构分析是研究基因功能的基础。有报道表明锌指结构在CESA之间的互作中起重要的作用,跨膜结构域是CESA形成三维结构的基础[3]。杨树次生壁CESA的结构分析发现,杨树和拟南芥次生壁CESA的锌指结构和跨膜结构的位置高度保守(图 1B)。杨树5个次生壁CESA的N端均含有锌指结构,杨树CESA4、CESA7A、CESA8A和CESA8B也与拟南芥的CESA4、CESA7和CESA8一样,均含有8个跨膜结构域,而杨树CESA7B的C端则缺少6个跨膜结构域。
-
基因表达分析研究有助于揭示基因的功能。为了获得杨树不同次生壁CESA基因的表达模式,检测了它们在不同组织及次生维管再生过程中的基因表达情况。杨树幼嫩与成熟的茎、叶和根组织的RNA-seq数据分析表明,杨树次生壁CESA基因在幼嫩叶、幼嫩茎、成熟茎以及根组织中均有可观的表达,但在成熟茎中表达量最高(图 2A)。值得注意的是,CESA8A在各个组织中的表达量均较其他几个CESA基因低(图 2A)。随后,通过对次生维管再生实验系统中不同时期再生组织基因表达的基因芯片数据分析表明,杨树次生壁CESA基因在剥皮12 d后的再生组织(木质部分化阶段)的表达量不断提高,与它们参与木材形成过程中次生壁纤维素合成的功能相吻合(图 2B)。另外,在这一过程中,5个杨树次生壁CESA基因的变化趋势一致,变化幅度相差不大。相对而言,CESA8A在再生组织中的表达量较其他几个CESA基因略低。
-
为了检测杨树次生壁CESA基因的组织表达特异性,创制了次生壁CESA基因启动子驱动GUS表达的转基因植株。通过GUS染色分析发现,在PPtCESA4: : GUS、PPtCESA7A: : GUS、PPtCESA8A: : GUS和PPtCESA7B: : GUS转基因植株的茎和叶中,均检测到较强的GUS信号(图 3)。在茎段中,顶端节间的GUS信号明显较基部节间的强。PPtCESA4: : GUS、PPtCESA7A: : GUS、PPtCESA8A: : GUS和PPtCESA7B: : GUS转基因植株之间,在叶中的GUS信号存在一定的差异。PPtCESA8A: : GUS和PPtCESA7B: : GUS转基因植株在整个叶片中均观测到了GUS信号(图 3B和3C),而PPtCESA4: : GUS和PPtCESA7A: : GUS转基因植株在叶脉中则基本上未检测到GUS信号(图 3A和3D)。
-
许多研究表明,激素可诱导CESA基因的表达[25-26]。利用赤霉素GA3、萘乙酸NAA、油菜素内酯BR、细胞分裂素6-BA和乙烯处理84K杨幼苗,检测了茎中次生壁CESA对激素的响应模式。结果显示,GA3和6-BA使杨树次生壁CESA的表达量上调,其中GA3的作用更显著(图 4A和4D)。NAA、BR和乙烯处理下,杨树次生壁CESA的表达量下调,其中BR的作用相对显著(图 4)。就5个杨树次生壁CESA基因的表达量变化对激素的响应而言,PtCESA4的表达量变化最显著,PtCESA7A较PtCESA7B的表达量变化显著,PtCESA8B较PtCESA8A的表达量变化显著。
-
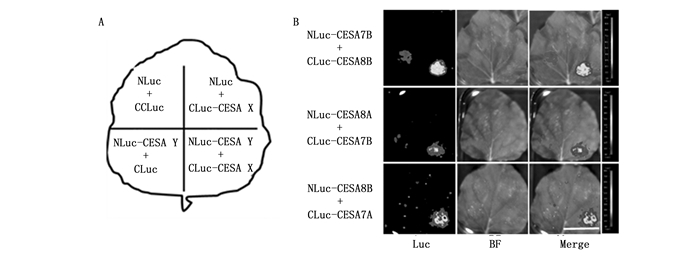
遗传学研究证实正常行使功能的CSC中含有3种CESA蛋白[27]。拟南芥中负责次生壁纤维素合成的CSC是由CESA4、CESA7和CESA8组成。在杨树中有5个次生壁CESA,其中1个CESA4、2个CESA7和2个CESA8。为了确定杨树中的2个CESA7和2个CESA8在与CESA4形成复合体的过程中究竟是几率均等还是有所偏好,对它们之间的互作情况进行了检测。如图 5所示,在荧光素酶互补报告体系中,相对于对照,PtCESA7B和PtCESA8B,PtCESA8A和PtCESA7B,PtCESA8B和PtCESA7A的组合表现出强的荧光信号,表明上述次生壁CESA可以互作形成二聚体,因此,PtCESA7A和PtCESA7B、PtCESA8A和PtCESA8B可能具有相同的能力形成有功能的CSC。
2.1. 杨树次生壁CESA基因的进化关系和蛋白结构
2.2. 杨树次生壁CESA在不同组织及次生维管发育中的表达特征
2.3. 基于启动子驱动GUS表达的转基因杨树中次生壁CESA的表达模式
2.4. 杨树次生壁CESA对激素处理的响应
2.5. 杨树次生壁CESA间的互作
-
纤维素合成需要纤维素合酶不同成员形成复合体,正常行使功能的CSC需要3种CESA蛋白[28]。本论文研究了杨树中5个次生壁纤维素合成相关的CESA基因的表达模式与互作模式,为揭示木本植物木材形成过程中5个次生壁CESA的作用提供了线索。研究结果表明,杨树5个次生壁CESA基因在杨树成熟茎中高表达,尤其在次生维管组织发育的后期高表达;CESA4、CESA7B、CESA8A和CESA8B的GUS信号在杨树茎和叶中较强,但这些次生壁CESA的表达模式存在一定的差异;杨树次生壁CESA的表达量被赤霉素和细胞分裂素上调,被生长素、油菜素内酯和乙烯下调;杨树次生壁CESA7B和CESA8B、CESA7B和CESA8A、CESA7A与CESA8B之间存在互作。这些数据提示杨树5个次生壁CESA基因虽然都能够参与形成CSC,但在不同组织、不同激素作用下可能有不同的组合方式。
参与次生壁纤维素合成的CSC需要由3种次生壁纤维素合成的CESA蛋白组成[15]。在拟南芥中,AtCESA4、AtCESA7与AtCESA8组成有功能的CSC,参与次生壁纤维素合成[13]。在水稻中,与AtCESA8、AtCESA4与AtCESA7同源的OsCESA4、OsCESA7与OsCESA9组成有功能的CSC,参与次生壁纤维素合成[29]。而在杨树中,有5个参与次生壁纤维素合成的CESA,其中包括了1个CESA4、2个CESA7和2个CESA8,它们有多达4种组合方式形成CSC,以参与次生壁纤维素的合成。RAN-seq与基因芯片的分析表明,杨树次生壁CESA基因在杨树成熟茎以及次生维管发育的后期即木质部形成时期表达量较高,提示它们在木材形成过程发挥重要作用。在这5个基因中,PtCESA8A的表达量略低,推测它被招募进入木材形成过程的CSC的机会略低。揭示杨树多个次生壁CESA以及多种次生壁纤维素合成CSC的存在,可为理解纤维素合成的质与量控制提供基础,进而为木材材性的多样性改良提供了潜在可能性。
基因结构分析发现,杨树5个次生壁CESA基因N端都具有锌指结构,为它们的互作提供了结构基础[15]。随后的荧光素酶互补实验也证实了这一点。CESA通常含有8个跨膜结构域,2个位于N端,6个位于C端[3]。然而,PtCESA7B的C端缺少6个跨膜结构域,不同于其它CESA,这种特殊的结构会否影响纤维素的合成目前尚不清楚。
杨树次生壁CESA启动子驱动GUS转基因杨树的GUS染色分析表明,这些基因的组织表达模式存在一定的差异。例如,PtCESA8A和PtCESA7B转基因株系整个叶片中均检测到了GUS信号,而PtCESA4和PtCESA7A转基因株系叶脉中则基本上未检测到GUS信号,此结果与Naoki等的研究结果一致[21]。这种差异提示在叶脉的发育中包含PtCESA7A的CSC作用应该大于包含PtCESA7B的CSC。另外,Naoki等的研究结果显示,PtCESA8B在木质部中高表达,而PtCESA8A则在根冠高表达;PtCESA7B在叶、叶柄、根冠和幼茎中表达量比PtCESA7A高,而PtCESA7A和PtCESA7B的表达量在成熟茎木质部中无明显差异[21]。这种组织表达的差异暗示,可能由不同的CESA组合形成不同的CSC,以参与不同组织器官的发育。
杨树次生壁CESA基因对不同激素处理的表达和响应存在较大差异。赤霉素GA3和细胞分裂素6-BA上调杨树次生壁CESA的表达,而NAA,BR和乙烯下调杨树次生壁CESA的表达。其中,PtCESA4基因的表达量变化最显著,PtCESA7A较PtCESA7B基因的表达量变化显著,PtCESA8B又较PtCESA8A基因的表达量变化显著。这一结果提示,由PtCESA4、PtCESA7A和PtCESA8B组成的CSC,在激素处理的响应中可能贡献更大。
荧光素酶互补实验结果表明,PtCESA7A、PtCESA7B、PtCESA8A和PtCESA8B之间可以相互作用。拟南芥次生壁CESA成员之间可以两两互作,形成二聚体,这种互作是其形成CSC的基础[15]。因此,PtCESA7A与PtCESA7B、PtCESA8A与PtCESA8B具有同等的几率形成有功能的CSC。然而,PtCESA7A与PtCESA7B、PtCESA8A与PtCESA8B在组织表达、激素响应上存在一定的差异,这些差异可能使得在不同组织、不同激素作用下,次生壁CSC招募不同的CESA成员参与次生壁纤维素的合成。CSC的多样性可以多方面影响细胞壁中的纤维素含量,从而导致木材材性差异。
-
杨树5种次生壁CESA基因具有同等的能力形成功能性的纤维素合酶复合体,但在不同组织、不同激素作用下可能有不同的组合方式。









 DownLoad:
DownLoad: