-
每个常规林木育种周期大体包括重组交配、遗传测定和根据育种值开展的前向或后向选择三部分[1-2],其中采用一定交配设计获得结构化系谱是后续遗传力和育种值等遗传参数估计的先决条件[3]。全双列是最完整、最能提供全面遗传信息的交配设计,但受限于时间和人力成本,亲本数量过多时难以实现。相反,自由授粉交配无需人工控制授粉、参试组合少、简便易行、成本低,是林木改良初期普遍采用的交配设计,但由于子代父本未知,仅能估计一般配合力(GCA),无法估计特殊配合力(SCA),同时由于存在近交、家系内个体并非纯半同胞[4-6]及父本贡献不均衡[7-10]等原因造成遗传参数估计不够准确,进而降低了育种效率。
采用DNA分子标记对自由授粉子代进行系谱重建不仅能够规避上述不足,而且可以矫正可能由人为原因造成的系谱错误[9, 11]。系谱重建最初由Lambeth等[12]提出,实现将多系交配的不完全谱系恢复为完全谱系,并将这些信息应用到单株线性混合模型中,除了准确地估算全部测定亲本及子代的育种值和非加性效应如亲本组合间的特殊配合力外,还能预测系谱恢复后未参与测定亲本的育种值[13]。系谱重建方法已被应用于桉属(Eucalyptus)[14]、海岸松(Pinus pinaster Ait.)[15]、花旗松(Pseudotsuga menziesii (Mirbel) Franco)[16]、云杉(Picea rubens Sarg.)[17]等多个林木改良计划中,采用的分子标记类型主要是简单重复序列(Simple Sequence Repeat,SSR),又叫微卫星标记,具有共显性、多态性高、稳定、易于开发等优点[18-20]。针对风媒授粉树种及其育种特性,El-Kassaby等[16]在系谱重建基础上提出了“免杂交育种(Breeding Without Breeding,BWB)”概念,旨在通过省去人工控制授粉和子代测定的环节达到缩短育种周期的目的,并利用该理念对花旗松商品林开展相关研究,将其育种周期缩短至4年,且能够获得正常育种的75%~85%的选择响应。类似地,Hansen和McKinney[21]使用12对SSR标记对高加索冷杉(Abies nordmanniana(Steven) Spach)商品林(种苗来源于种子园内99个无性系)及其亲本进行分型,将系谱重建后的数据作为类似子代测定的数据计算相关遗传参数并加以选择,达到缩短育种周期的目的。然而该理念存在的一些短板限制了它在实际育种中的应用,例如对于雌雄同株树种,商品林个体的系谱重建只能鉴定出亲本而无法区分父母本,而在种子园存在花粉污染的情况下,更增加了商品林中子代亲本的甄别难度和分型成本。此外,对于亲子关系可信度较低及鉴定到一个亲本的个体,在采用全同胞模型分析时无法利用全部信息[22],同时种子园亲本生殖力存在较大差异,大部分自由授粉子代可能来自于少数父本,造成贡献率低的父本的育种值估计不准确,而且通常高父本贡献产生的子代的生长表现较为一般[23]。因此,系谱重建或BWB策略更适用于那些基础薄弱的树种或初期营建的自由授粉子代测定林等,而对于高世代遗传改良项目则必须采用更为科学而严谨的交配设计和遗传测定[3]。
日本落叶松(Larix kaempferi(Lamb.) Carr.)是我国引进的主要针叶纸浆用材树种[24],具有早期速生、成林快、易于栽培、适应性广等特点[25]。本研究以1988年营建于辽宁省大孤家林场的日本落叶松自由授粉子代测定林为材料,利用课题组前期开发的SSR标记[23],对来自66个自由授粉家系的601个子代进行系谱重建,研究系谱重建前后生长和化学材性性状遗传参数的变化,以期为国内以自由授粉子代测定林为主的遗传测定提供更为科学有效的评价方法。
HTML
-
以1988年建于辽宁大孤家林场(42.72°N,124.88°E,海拔约460 m)的子代测定林为对象。试验林采用随机完全区组设计,共包含96个自由授粉家系,5次重复,小区株数为5~15株不等,2年生苗木造林,株行距为2 m×2 m。选择其中保存株数较多的66个家系,每个家系随机选择10~11个子代,共计668个单株组成试验群体。子代测定林家系种子采自该场1965年营建的日本落叶松无性系种子园,建园优树选自辽宁本溪、草河口、抚顺、庄河等地30~40年生早期引种的日本落叶松人工林林分,优树选择方法采用3株或5株大树法,采用人工嫁接方法建园。1985年采种,当时种子园内无性系140个,无性系采取错位配置,确保同一无性系间距不小于24 m。
-
于2013年采集668个子代及种子园亲本针叶用于DNA提取,采用17对EST-SSR标记和毛细管电泳进行基因分型。利用CERVUS 3.0[26]软件进行父本分析,基于LOD得分确定子代的父本。候选父本的数量设为140,候选父本的比例和分型错误率分别设为0.85和0.01,最少位点数设为9。在严格(95%)和宽松(80%)两个置信水平上分别模拟1万次,最终有601个子代成功恢复父本信息,单一亲本基因型已知时的联合排除概率大于99.9%[23]。
-
2005年12月对测定林开展每木生长调查(17年生)。树高(H,m)采用激光测高仪(Vertex Ⅲ,Haglof Company Group,Sweden)测定,胸径(DBH,cm)采用测径尺测定,并计算材积指数(V,dm3):
化学材性测定采用快速高效的无损检测方法-近红外光谱(Near Infrared Reflectance,NIR)扫描法[27-28]。大致流程为在每株树胸高处用生长锥钻取直径为5 mm的木芯,室内气干后研磨过40目筛,NIR扫谱后利用课题组建立的高精度化学材性预测模型预测α-纤维素、木质素和综纤维素含量[23]。
-
利用单株线性混合模型在R软件(版本3.1.2)ASReml-R包(版本3.0)中限制性最大似然估计方法(REML)估计随机效应的方差分量[29-30]。利用似然比检验(likelihood ratio test,LRT)各方差组分的统计显著性。利用泰勒级数展开法(Taylor series expansion)计算遗传参数的标准误[29]。对于系谱重建前的自由授粉家系,假定显性和上位效应不存在,采用基础模型(B)计算:
其中,y是观测值向量,β是固定的区组效应,a和p分别是随机的加性和小区效应向量,e是随机残差;X、Z1和Z2分别是对应效应的关联矩阵。
系谱重建后,假定上位和正反交效应为零,采用系谱重建模型(PR)计算:
其中,f是家系效应(即特殊配合力),Z3是家系效应的关联矩阵,其余同模型(2)。
模型(2)和(3)相应的遗传力计算公式分别为:
其中,$\hat \sigma _d^2 = 4 \times \hat \sigma _f^2, \hat \sigma _a^2, \hat \sigma _d^2, \hat \sigma _f^2、\hat \sigma _p^2$和$\hat \sigma _e^2$分别是加性方差、显性方差、家系方差、小区方差和残差方差的估计值。
通过BLUP(最佳线性无偏预测)法求解混合模型方程组即可得到随机效应值[31]。混合模型方程组的一般形式为:
其中,X=⊕i-1m Xi,Z=⊕j-1n Zj,m和n分别是固定和随机效应关联矩阵的个数;A是加性关系矩阵;$ \alpha=(1-\hat{h})^{2} / \hat{h}^{2}$;$\hat{b}$和$\hat{u}$分别是固定和随机效应向量。
公式(5)中等号左侧系数矩阵可以写作:$\left[{array}{cc}{\text{C}_{11}} & {\text{C}_{12}} \\ {\text{C}_{21}} & {\text{C}_{22}}{array}\right]$,其广义逆矩阵为$\left[{array}{cc}{\text{C}^{11}} & {\text{C}^{12}} \\ {\text{C}^{21}} & {\text{C}^{22}}{array}\right]$。
预测育种值误差方差(Prediction Error Variance,PEV)是育种值的标准误的平方和[32],计算公式为:
其中a为加性效应值的真值。
然后,育种值的准确度可以由下式得出:
亲本和子代的育种值准确度分别用rp和ro表示。
采用斯皮尔曼秩相关(Spearman rank correlation)分析并检验系谱重建前后育种值排名的相关性。采用基于BLUP估计的育种值计算前向选择的遗传增益(G)[33-34],入选率设为5%。
1.1. 试验材料
1.2. 系谱重建
1.3. 表型数据
1.4. 数据分析
-
各性状的表型统计见表 1。601株日本落叶松19a生树高和胸径的变幅分别为10.5~24.0 m和9.1~24.3 cm,胸径的变异系数略高于树高(14.42% vs. 11.74%)。α-纤维素、木质素和综纤维素含量的均值分别为45.37%、28.11%和72.40%,其中α-纤维素含量的变异系数最大(9.43%)。化学性状的变异系数均小于生长性状。
性状
Trait均值
mean标准差
sd极小值
min极大值
max表型变异系数CV/% 树高tree height/m 17.59 2.06 10.50 24.00 11.74 胸径diameter at breast height/cm 16.55 2.39 9.10 24.30 14.42 材积指数
volume index/dm350.16 18.46 10.35 115.82 36.79 α-纤维素
α-cellulose/%45.37 4.28 33.31 57.06 9.43 木质素lignin/% 28.11 1.53 23.41 33.48 5.44 综纤维素
holo-cellulose/%72.40 4.34 59.65 86.39 5.99 Table 1. Descriptive statistics for growth and wood chemical traits
-
采用经典的单株混合模型分别对自由授粉家系和系谱重建后的全同胞家系数据进行比较分析,各效应方差分量及遗传力列于表 2。系谱重建后PR模型预估的各生长性状的加性方差(${\hat \sigma _a^2}$)远低于基础模型B;同时PR模型的遗传力也从B模型的中高度遗传(${{\hat h}^2}$>0.3)降至中低度遗传(${{\hat h}^2}$ = 0.1,胸径和材积指数),树高的遗传力甚至接近于0,说明B模型高估了遗传力;PR模型中${\hat \sigma _a^2}$和${{\hat h}^2}$的标准误均小于B模型,说明其预估精度更优;树高主要受显性效应控制,胸径和材积则主要受加性效应控制。B模型中材性性状的小区方差(${\hat \sigma _p^2}$)均不显著,残差方差接近于0,${\hat \sigma _a^2}$几乎解释了全部变异;系谱重建后${\hat \sigma _a^2}$有所下降而${\hat \sigma _p^2}$显著增加,说明自由授粉家系忽略了全同胞家系与区组间交互作用这一事实;${{\hat h}^2}$的估计值更趋于合理,木质素遗传力最高(${{\hat h}^2}$ = 0.55),α-纤维素最低(${{\hat h}^2}$ = 0.35)。α-纤维素和木质素主要受加性效应控制,同时显性效应也作用显著。因此,通过系谱重建补全父本信息后,B模型中的大部分${\hat \sigma _a^2}$被分流到PR模型的${\hat \sigma _d^2}$, 从而提高了遗传参数的预估精度,同时因为PR模型含有更多的遗传信息,增加了分析的敏感性,模型中${\hat \sigma _p^2}$和${\hat \sigma _e^2}$虽略有升高,而标准误则更低。
模型
Model性状
Trait加性方差
${\hat \sigma _a^2}$(se)显性方差
${\hat \sigma _d^2}$(se)小区方差
${\hat \sigma _p^2}$(se)残差方差
${\hat \sigma _e^2}$(se)遗传力
${{\hat h}^2}$(se)亲本育种值准确度rp 子代育种值准确度ro 遗传增益G B模型 树高
tree height/m1.15
(0.55)NA 0.94
(0.21)1.08
(0.44)0.36
(0.16)0.58 0.51 3.09 胸径diameter at breast height/cm 1.38
(0.65)NA 0.19
(0.28)3.05
(0.6)0.3
(0.14)0.60 0.55 3.69 材积指数
volume index/dm375.49
(36.73)NA 11.35
(15.82)176.78
(33.62)0.29
(0.14)0.55 0.51 8.94 α-纤维素
α-cellulose/%9.79
(0.87)NA 1.05
(0.66)ns<0.001
(<0.001)0.90
(0.06)0.68 0.67 18.33 木质素
lignin/%1.81
(0.1)NA 0.01
(<0.001)ns<0.001
(<0.001)1
(<0.001)0.71 0.77 5.96 综纤维素
holo-cellulose/%10.82
(0.95)NA 0.89
(0.7)ns<0.001
(<0.001)0.92
(0.06)0.70 0.76 17.76 PR模型 树高
tree height/m0.02
(0.16)ns0.45
(0.25)1.26
(0.21)1.5
(0.26)0.006
(0.015)NA NA NA 胸径diameter at breast height/cm 0.65
(0.28)0.06
(0.4)ns0.33
(0.28)3.59
(0.5)0.14
(0.06)0.72 0.62 2.72 材积指数
volume index/dm327.65
(11.77)0.82
(1.13)ns22.28
(15.01)212.91
(27.37)0.11
(0.07)0.68 0.57 5.38 α-纤维素
α-cellulose/%5.64
(0.44)1.46
(0.66)8.79
(1.15)<0.001
(<0.001)0.35
(0.06)0.81 0.91 8.06 木质素
lignin/%1.24
(0.07)0.47
(0.11)0.48
(0.13)0.06
(0.02)0.55
(0.04)0.84 0.94 3.48 综纤维素
holo-cellulose/%7.55
(0.8)1
(0.6)ns7.74
(1.17)0.56
(0.34)0.45
(0.03)0.83 0.92 8.75 注: NA—缺失no available, 下同;se-标准误standard error; ns—不显著not significant. Table 2. Comparison of estimates of variance components, breeding value accuracy and realized genetic gain from B and PR model
-
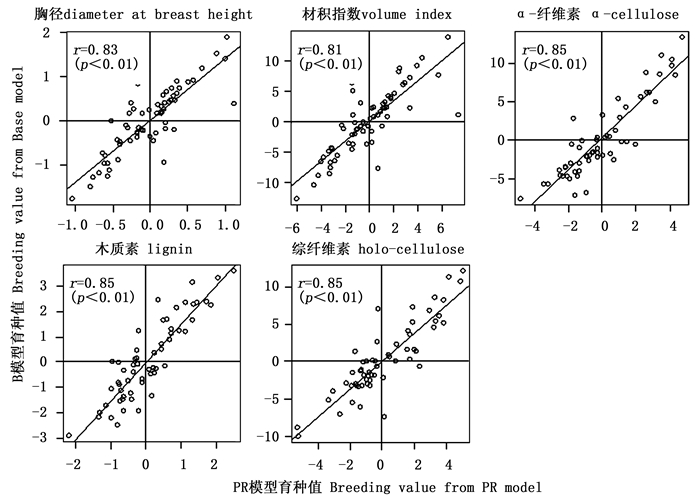
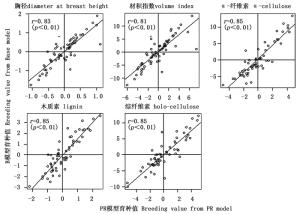
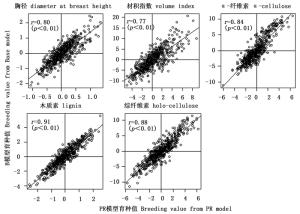
由于树高在PR模型的加性方差接近于零,相应的育种值也极小,因此后续分析中未考虑该性状。通过比较2个模型预估的育种值相关性发现,无论是亲本育种值,还是子代育种值2个模型间均存在着极显著的一致性(r≈0.83,p<0.01)(图 1、2),其中材性的相关性较生长性状的相关性更高,尤其是木质素和综纤维素(r≈0.9)。按10%的选择强度,系谱重建前后对亲本进行后向选择时(7株)各性状存在14%~29%的选择差异,对子代进行前向选择时(30株)胸径、材积和α-纤维素存在30%~35%的选择偏差,木质素和综纤维素分别存在7%和19%的选择偏差。
系谱重建后生长性状的亲本育种值准确度较系谱重建前提高了20%~24%,子代育种值准确度提高了12%~13%;系谱重建后材性性状的亲本育种值准确度提高了18%~19%,子代育种值准确度提高了21%~35%(表 2)。总体而言,材性性状的育种值准确度普遍高于生长性状的育种值准确度,同一生长性状的亲本育种值准确度普遍高于子代育种值准确度,完全系谱PR模型预估的育种值准确度均高于自由授粉家系B模型预估的育种值准确度。
-
现实遗传增益随着系谱重建后${\hat \sigma _a^2}$的降低而下降,当以5%的强度进行前向选择时,胸径和材积的现实遗传增益分别为2.72%、5.38%,降幅达26.3%和39.8%;α-纤维素、木质素(含量低者为优)及综纤维素含量的遗传增益,分别为8.06%、3.48%和8.75%,降幅分别为56%、42%和51%(表 2)。材性性状的遗传增益普遍高于生长性状,说明该试验林材性性状具有更大的改良潜力。
2.1. 表型描述
2.2. 方差组分和遗传力
2.3. 育种值
2.4. 遗传增益
-
常规林木遗传改良的长周期性决定了在育种实践中需要投入大量的人力、物力和时间成本,因此制定合适的育种策略并提高育种效率显得尤为重要。加性(或显性)方差、遗传力及育种值等遗传参数的准确估算是育种策略制定的依据,而育种效率的提升则主要依赖于合理的交配设计、田间试验和统计分析方法等。自由授粉家系子代测定因其简便易行是林木改良初期交配设计的首选,但早在上世纪六七十年代林木育种学家就意识到,该交配设计由于无法满足一些基本的遗传和统计假设,而导致遗传参数估计的偏差[4-5],并尝试采用多系混合授粉[8, 35]或种子园人工辅助授粉[36-37]等方式改进自由授粉交配的不足,但同样存在无法区分子代父本的缺陷。同时由于近交的存在,在部分树种中证实采用加性遗传方差等于2.5倍家系方差(或计算得到的矫正系数)而非4倍家系方差计算的遗传力更准确[38-39],但该方法仍然无法满足不同树种乃至不同群体亲本间自由交配的假设,也无法校正育种值被高估的错误。而基于DNA分子标记的系谱重建技术为自由授粉测定材料的遗传参数的准确估计提供了新的思路。
本研究比较了来自66个自由授粉家系的601个子代群体系谱重建前后生长及材性性状的方差分量、遗传力和育种值等遗传参数,以及育种值准确度和遗传增益的变化。研究结果显示,由于常规的自由授粉家系B模型无法区分显性方差${\hat \sigma _d^2}$分量,混在估计的加性遗传方差${\hat \sigma _a^2}$中,因此会高估${\hat \sigma _a^2}$和遗传力,这在多个报道中也同样得到证实[9, 17, 22, 40]。树高的${\hat \sigma _a^2}$在系谱重建后从1.15降至近乎为0,这可能与子代父本的贡献极不均衡有关[23],系谱重建后家系内个体数量变化很大,部分家系只有1个子代,而树高可能对样本量的变化更为敏感。树高主要受显性效应控制,胸径和材积主要受加性效应控制,材性的化学性状除了受加性效应控制外,也受显性效应控制。如果仅是高估遗传力而不影响育种值排名,则不会影响遗传改良效率。系谱重建方法能够显著提高育种值估算的准确度,从而更加准确地预估现实遗传增益,尤其是当测定群体数量较大时更为重要。除此之外,系谱重建后还能够估算重建父本的育种值,当样本量足够大时就能对种子园所有交配亲本进行育种值估算和选择。
系谱重建在林木遗传改良的应用中具有多方面的优势。一方面可以提供种子园交配模式、父本贡献和花粉污染等的有效信息,有助于提高种子园的经营管理;另一方面可以恢复子代的父本信息,有助于更准确地估算育种值等遗传参数,同时可以减少新入选子代间的亲缘关系,进一步为育种策略的制定提供科学依据。系谱重建策略是解决既有自由授粉子代遗传测定缺陷的有效方法,但也应充分认识到系谱重建在应用中的不足。为了鉴定出更多的亲子关系,通常采用80%的置信度进行分析[41],导致约5%的错误率[41-42],因此为了减少错误率应尽可能提高置信度的阈值。此外,在系谱重建前要先排查是否存在无效等位基因(null allele)和突变,否则也会导致父本的排除错误。单株模型依赖于个体间的亲缘关系系数,无论是半同胞还是全同胞均假定同胞间有相同的关系系数,然而这一假设在现实中是不存在的[5],因此即便能够全部重建自由授粉子代的父本信息,采用的依然是理论上的亲缘关系系数。DNA分子标记也可以用来估计个体间的亲缘关系,并且可以利用历史遗传信息[43],当标记数量过少时获得的结果并不准确[44-45],只有基于大量分子标记获得的亲缘关系可以替代系谱关系[46],但由于其高昂的成本对于多数林木遗传改良项目,目前还难以承受。一般情况下系谱重建很难识别全部子代的亲本从而造成数据缺失,这在利用BWB方法对商品林中的个体进行系谱重建时表现得更为突出。此外,种子园父本间的贡献率存在较大的差异,即父本贡献的非均衡性,导致大部分自由授粉子代来自于少数父本,而且通常最多父本贡献的子代的生长表现一般[23]。因此,对于当前国内的自由授粉测定材料,建议采用基于单株模型的系谱重建或系谱重建结合少量标记方法取代常规基于亲本模型的分析方法,从而获得更为准确的遗传参数、提高育种效率。
对于进入高世代的遗传改良项目则必须转而采用更加科学而严谨的交配设计。当育种群体数量较大时,如何对其开展有效的交配既保证遗传增益的最大化同时又保持一定的遗传多样性?半不连续双列交配设计是火炬松、辐射松、花旗松等高世代育种项目中主要采用的交配设计[47-49]。为了控制育种群体中的近交水平,Burdon和Namkoong[50]提出了“亚系”(subline)的概念,将育种群体划分成多个组(亚系),从育种角度交配和选择只允许在亚系内进行,而从生产角度则是在亚系间进行。该策略已应用在多个高世代改良项目中,能较好地维持育种群体的遗传多样性,但缺点是管理成本较高且损失部分遗传增益[51]。最近,Kinghorn[52]提出了利用线性规划寻找最佳交配组合的一种有效方法,并开发出了“Pedigree view”的计算机软件,能够同时解决交配组合选择与近交控制等问题,实现育种群体的遗传增益和遗传多样性的平衡。
-
系谱重建能够恢复自由授粉子代的系谱信息,而完整的系谱信息又能够获得更为丰富的遗传信息及更加准确的遗传参数和育种值,有助于提高自由授粉子代测定的选择效率及优化育种策略。系谱重建策略是解决国内既有自由授粉子代遗传测定缺陷的行之有效的方法,但对于进入高世代的遗传改良项目则必须采用更加科学合理的控制授粉交配设计和遗传测定。



 DownLoad:
DownLoad:
